 |
 |
 |
The project
- Acute Lymphoblastic Leukemia detection based on adaptive unsharpening and Deep Learning: [GitHub]
- Histopathological transfer learning for Acute Lymphoblastic Leukemia detection: [GitHub]
- ALLNet: Acute Lymphoblastic Leukemia detection using lightweight convolutional networks: [GitHub]
- ALL-IDB Patches: Whole slide imaging for Acute Lymphoblastic Leukemia detection using Deep Learning: [GitHub]
- DL4ALL: Multi-task cross-dataset transfer learning for Acute Lymphoblastic Leukemia detection: [GitHub]
- A decision support system for Acute Lymphoblastic Leukemia detection based on Explainable Artificial Intelligence: [GitHub]
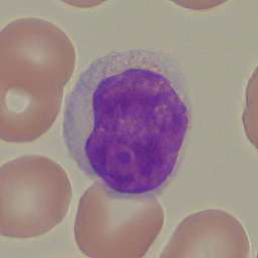
Acute Lymphoblastic (or Lymphocytic) Leukemia (ALL) is a disease that affects the blood cells, can spread rapidly throughout the body, and may result in fatal consequences if not detected at an early stage. One of the techniques routinely used to diagnose ALL consists in analyzing White Blood Cells (WBC) present in peripheral blood samples to look for malformations or abnormalities. Such malformations may be an indicator of lymphoblasts, which naturally occur in the bone marrow. However, an elevated number of WBCs with lymphoblast characteristics may be a sign of ALL.
Traditionally, the analysis of the WBC morphology is performed manually by an expert pathologist, who looks at the blood cells and estimates the concentration of lymphoblasts present in peripheral blood. Such process, being extremely repetitive and time-consuming, may lead to fatigue, with the consequence that the pathologist could miss important information correlated with the presence of ALL.
To overcome the disadvantages of a manual inspection process, Computer Aided Diagnosis (CAD) systems are being increasingly researched: such systems are often based on image processing and machine learning and, by automatically detecting lymphoblasts, can help the pathologist in performing a preliminary screening of the blood samples. Among CAD systems, recent methods are increasingly considering the use of machine learning approaches based on Deep Learning (DL) and Convolutional Neural Networks (CNN), due to their high accuracy in several fields, including medical imaging. In particular, CNNs have the ability of automatically learning data representations, without the need for a handcrafted feature extraction step, with the consequence that CAD systems based on CNNs may be designed with limited knowledge of the application scenario.
The method
Acute Lymphoblastic Leukemia detection based on adaptive unsharpening and Deep Learning
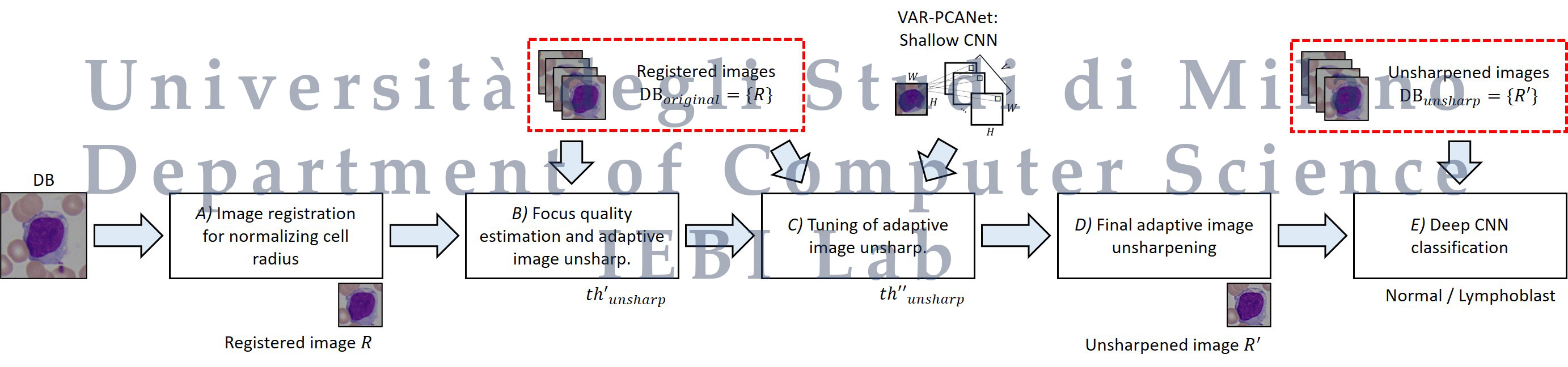
We propose the first machine learning-based approach able to enhance blood sample images by an adaptive unsharpening method. The method uses image processing techniques and DL to normalize the radius of the cell, estimate the focus quality, adaptively improve the sharpness of the images, and then perform the classification. We evaluated the methodology on a public database of ALL images, considering several state-of-the-art CNNs to perform the classification, with results showing the validity of the proposed approach.
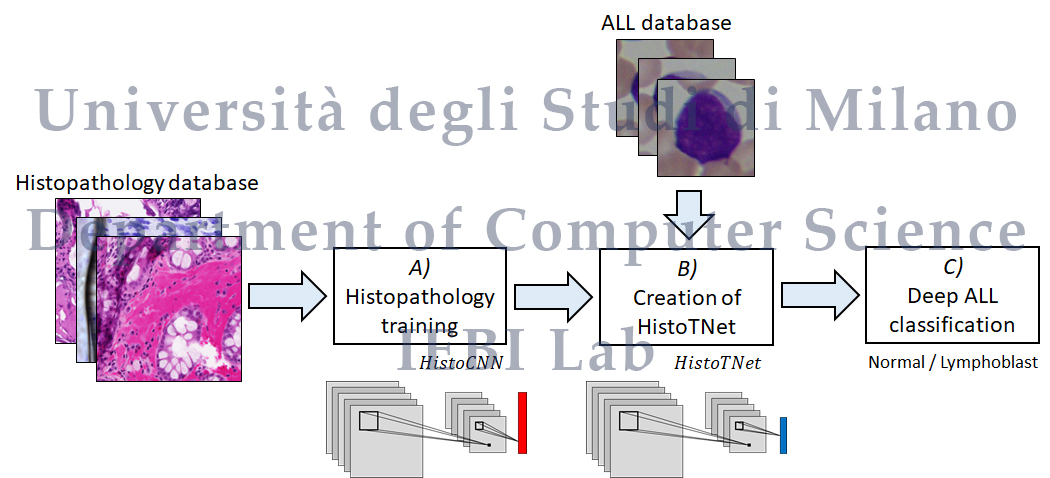
Histopathological transfer learning for Acute Lymphoblastic Leukemia detection
We propose the first method based on histopathological transfer learning for ALL detection, which trains a CNN on a histopathology database to classify tissue types, then performs a fine tuning on the ALL database to detect the presence of lymphoblasts. As histopathology database, we consider a multi-label dataset with a significantly higher number of samples and classes with respect to the literature, which enables CNNs to learn general features for histopathology image processing and hence allow to perform a more effective transfer learning, with respect to CNNs pretrained on ImageNet. We evaluate the methodology on a publicly-available ALL database and considering multiple CNNs, with results confirming the validity of our approach.
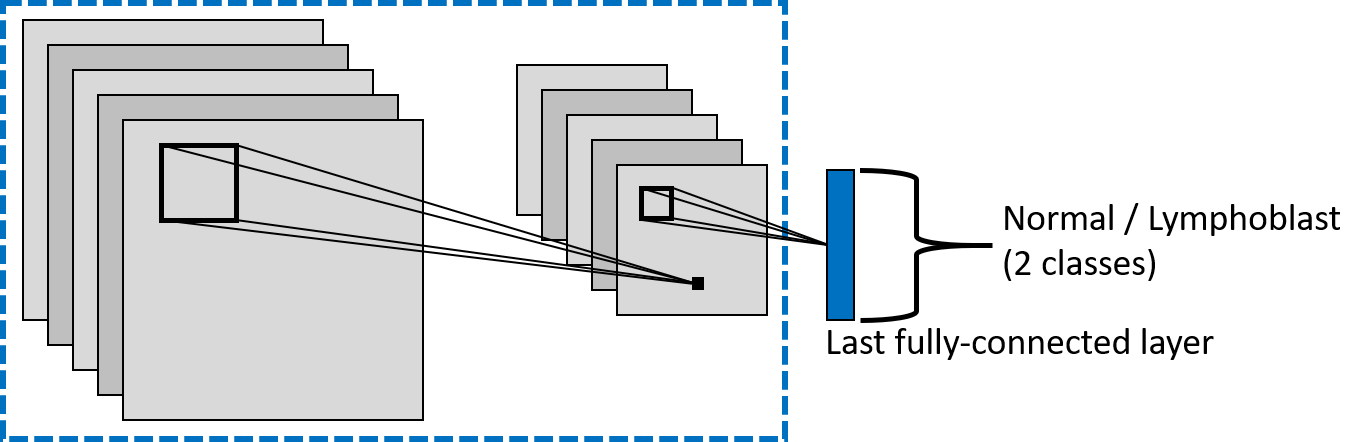
ALLNet: Acute Lymphoblastic Leukemia Detection Using Lightweight Convolutional Networks
We propose ALLNet, the first approach in the literature for ALL detection using a lightweight architecture based on fixed binary kernels that replicate the Local Binary Patterns and that uses only ≈1.6% of the parameters of a traditional CNN, at the same time achieving better results in terms of classification accuracy.
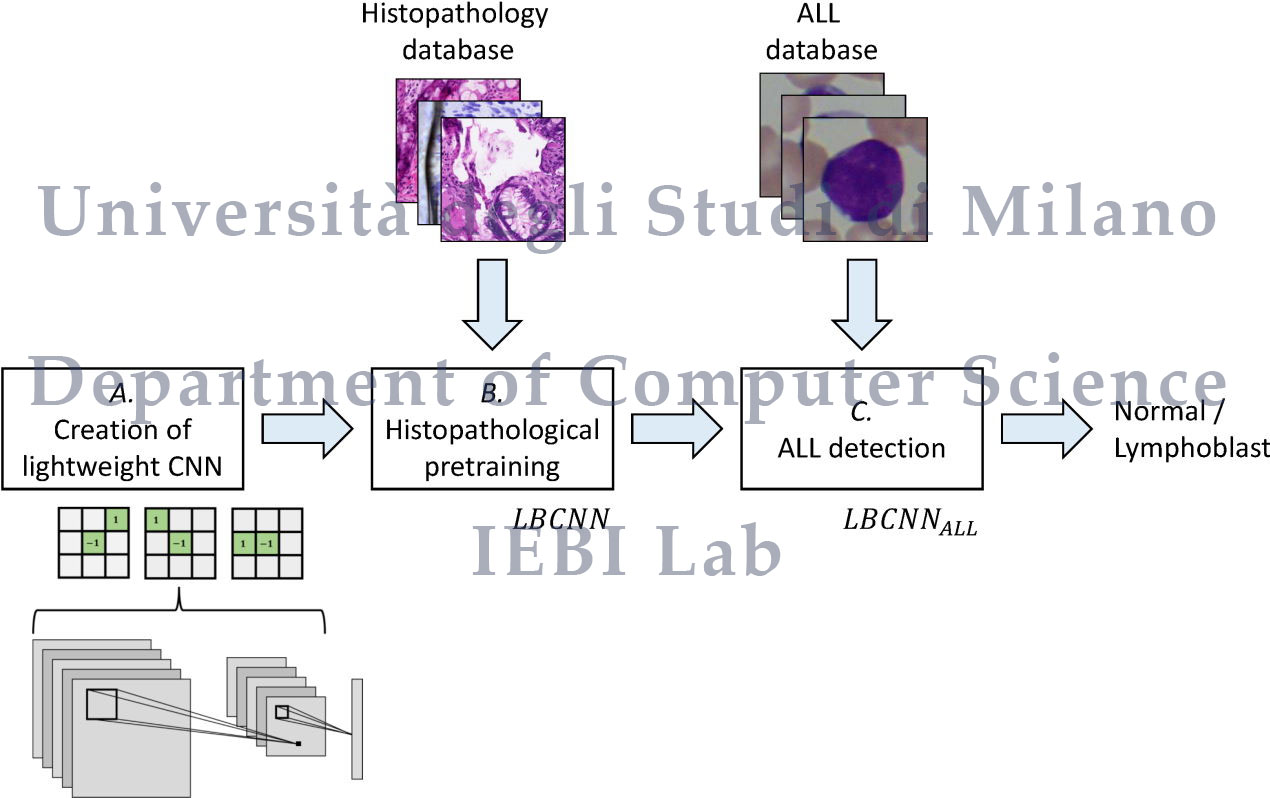
Structure of the models used in the paper (PyTorch):
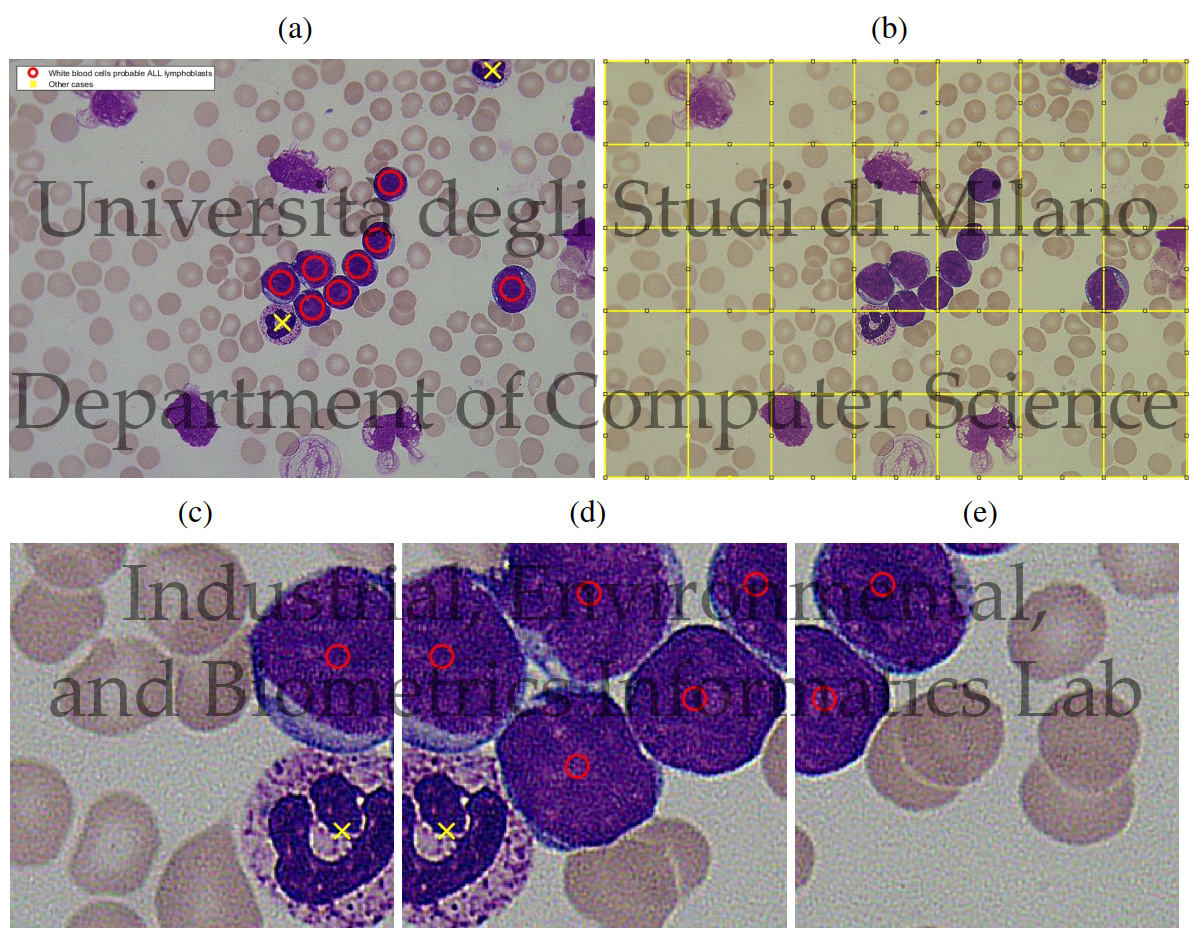
ALL-IDB Patches: Whole slide imaging for Acute Lymphoblastic Leukemia detection using Deep Learning
We propose the ALL-IDB Patches approach, which consists in cropping portions of the WSIs contained in the ALL-IDB1 dataset, with the purpose of making the WSIs usable for DL-based algorithms and exploiting all the information contained in WSIs.
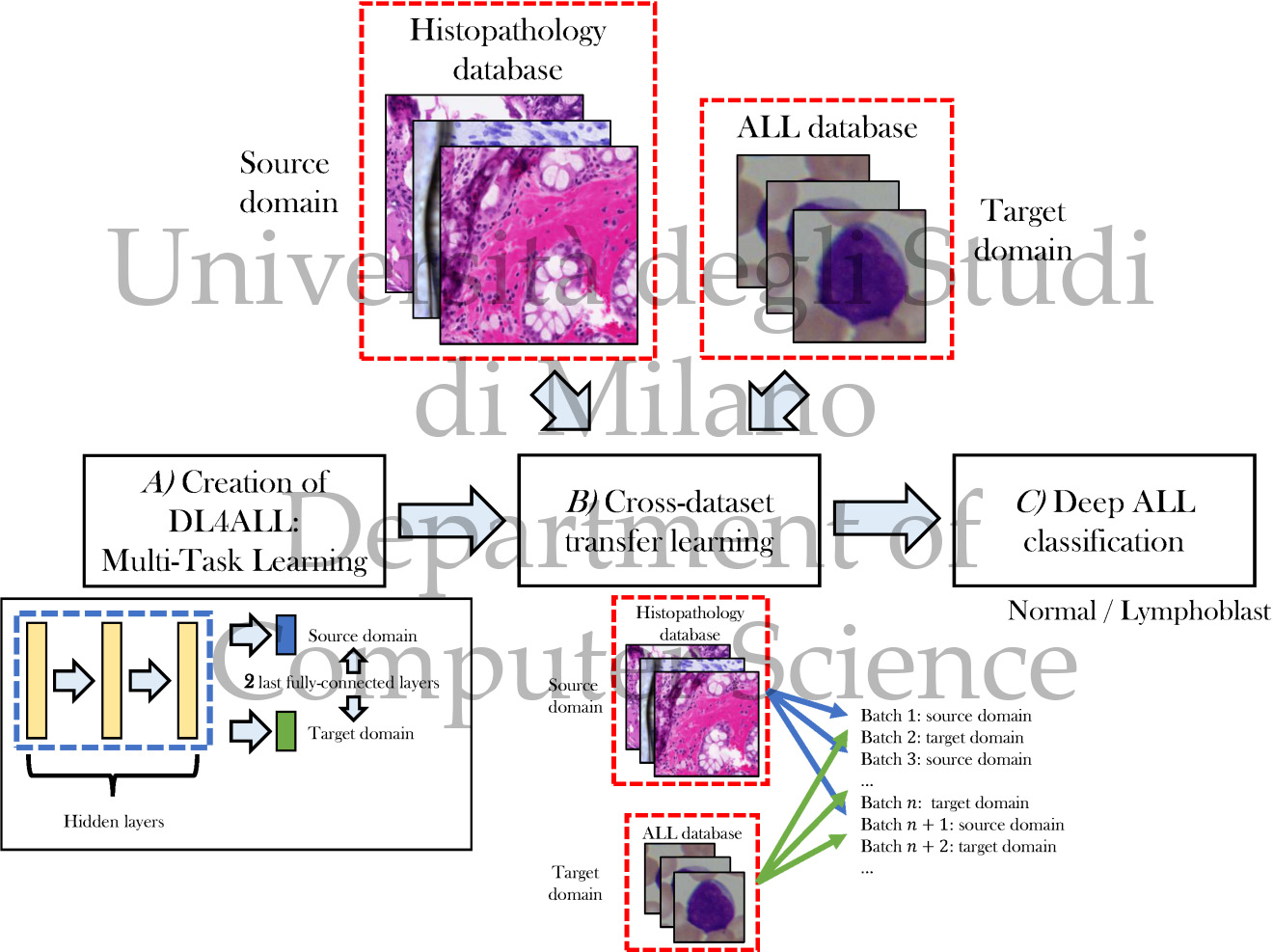
DL4ALL: Multi-task cross-dataset transfer learning for Acute Lymphoblastic Leukemia detection
We propose "Deep Learning for Acute Lymphoblastic Leukemia" (DL4ALL), a novel multi-task learning DL model for ALL detection, trained using a cross-dataset transfer learning approach. The method adapts an existing model into a multi-task classification problem, then trains it using transfer learning procedures that consider both source and target databases at the same time, interleaving batches from the two domains even when they are significantly different.
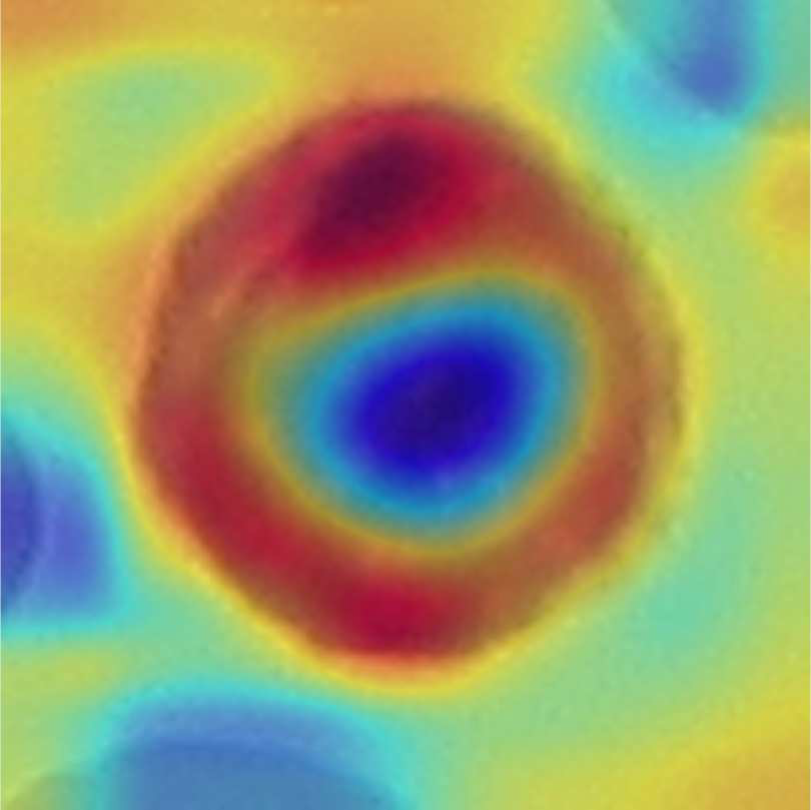
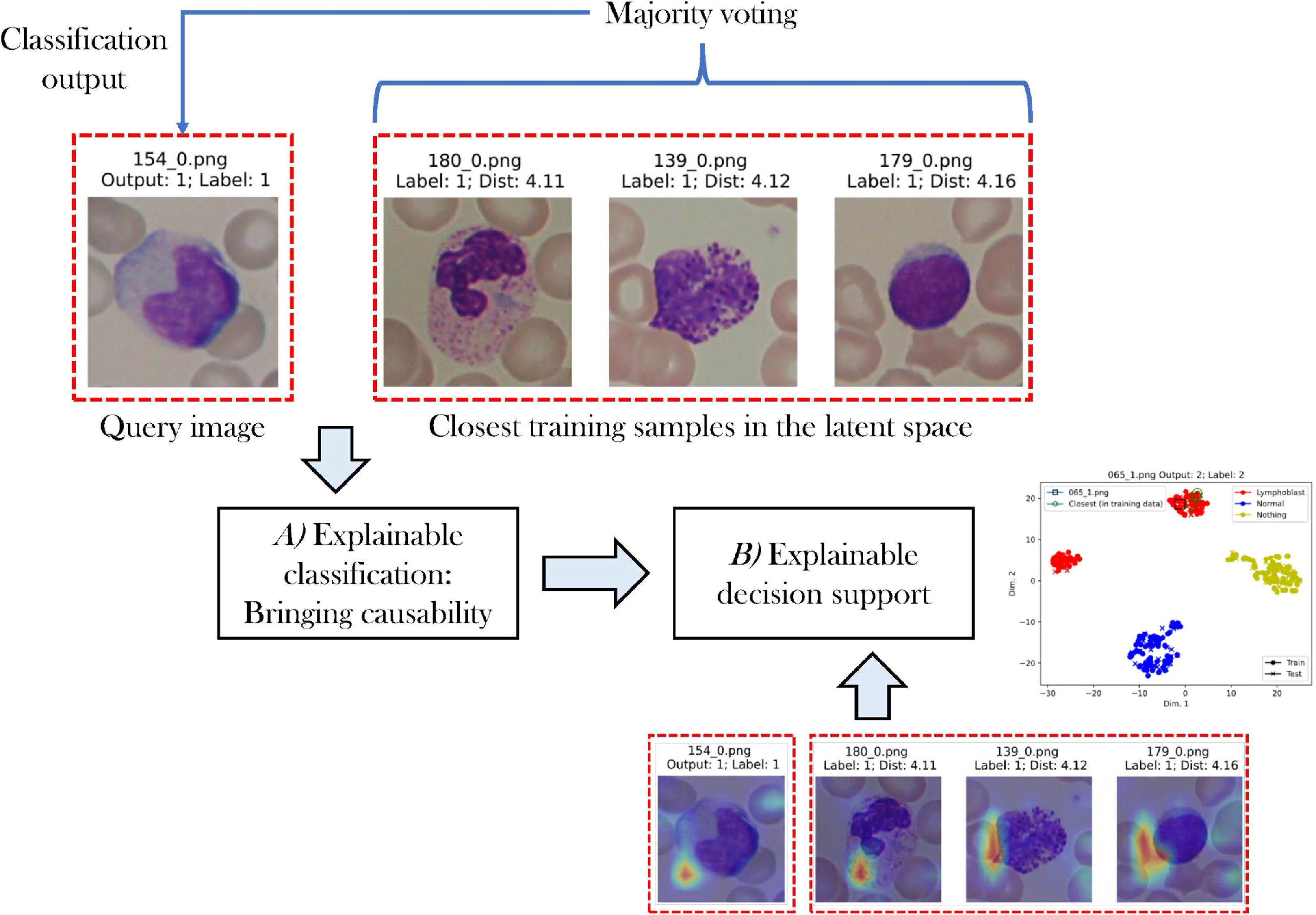
A decision support system for Acute Lymphoblastic Leukemia Detection based on Explainable Artificial Intelligence
We propose an innovative decision support system for ALL detection that is based on DL and explainable artificial intelligence (XAI). Our approach first introduces causality into the decision with a metric learning approach, enabling a decision to be made by analyzing the most similar images in the database. Second, our method integrates XAI techniques to allow even non-trained personnel to obtain an informed decision by analyzing which regions of the images are most similar and how the samples are organized in the latent space.
Related publications
- A. Genovese, M. S. Hosseini, V. Piuri, K. N. Plataniotis, and F. Scotti, "Acute Lymphoblastic Leukemia detection based on adaptive unsharpening and Deep Learning", in Proc. of the 2021 IEEE Int. Conf. on Acoustics, Speech, and Signal Processing (ICASSP 2021), Toronto, ON, Canada, June 6-11, 2021, pp. 1205-1209. ISBN: 978-1-7281-7605-5. [DOI: 10.1109/ICASSP39728.2021.9414362] [PDF] [BibTex entry]
- A. Genovese, M. S. Hosseini, V. Piuri, K. N. Plataniotis, and F. Scotti, "Histopathological transfer learning for Acute Lymphoblastic Leukemia detection", in Proc. of the 2021 IEEE Int. Conf. on Computational Intelligence and Virtual Environments for Measurement Systems and Applications (CIVEMSA 2021), June 18-20, 2021, pp. 1-6. ISBN: 978-1-6654-1249-0. [DOI: 10.1109/CIVEMSA52099.2021.9493677] [PDF] [BibTex entry]
- A. Genovese, "ALLNet: Acute Lymphoblastic Leukemia detection using lightweight convolutional networks", in Proc. of the 2022 IEEE Int. Conf. on Computational Intelligence and Virtual Environments for Measurement Systems and Applications (CIVEMSA 2022), Chemnitz, Germany, June 15-17, 2022, pp. 1-6. ISBN: 978-1-6654-3445-4. [DOI: 10.1109/CIVEMSA53371.2022.9853691] [PDF] [BibTex entry]
- A. Genovese, V. Piuri, and F. Scotti, "ALL-IDB Patches: Whole slide imaging for Acute Lymphoblastic Leukemia detection using Deep Learning", in Proc. of the IEEE Int. Conf. on Acoustics Speech and Signal Processing Workshops (ICASSPW 2023), Rhodes Island, Greece, June 4-10, 2023, pp. 1-5. ISBN: 979-8-3503-0261-5. [DOI: 10.1109/ICASSPW59220.2023.10193429] [PDF] [BibTex entry]
- A. Genovese, V. Piuri, K. N. Plataniotis, and F. Scotti, "DL4ALL: Multi-task cross-dataset transfer learning for Acute Lymphoblastic Leukemia detection", in IEEE Access, vol. 11, 2023, pp. 65222-65237. ISSN: 2169-3536. [DOI: 10.1109/ACCESS.2023.3289219] [PDF] [BibTex entry]
- A. Genovese, V. Piuri, and F. Scotti, "A Decision Support System for Acute Lymphoblastic Leukemia Detection based on Explainable Artificial Intelligence", in Image and Vision Computing, vol. 151, no. 105298, November, 2024. ISSN: 0262-8856. [DOI: 10.1016/j.imavis.2024.105298] [PDF] [BibTex entry]
Citations
@InProceedings {icassp21,
author = {A. Genovese and M. S. Hosseini and V. Piuri and K. N. Plataniotis and F. Scotti},
booktitle = {Proc. of the 2021 IEEE Int. Conf. on Acoustics, Speech, and Signal Processing (ICASSP 2021)},
title = {Acute Lymphoblastic Leukemia detection based on adaptive unsharpening and Deep Learning},
address = {Toronto, ON, Canada},
pages = {1205-1209},
month = {June},
day = {6-11},
year = {2021},
note = {978-1-7281-7605-5},
}
@InProceedings {civemsa21all,
author = {A. Genovese and M. S. Hosseini and V. Piuri and K. N. Plataniotis and F. Scotti},
booktitle = {Proc. of the 2021 IEEE Int. Conf. on Computational Intelligence and Virtual Environments for
Measurement Systems and Applications (CIVEMSA 2021)},
title = {Histopathological transfer learning for Acute Lymphoblastic Leukemia detection},
pages = {1-6},
month = {June},
day = {18-20},
year = {2021},
note = {978-1-6654-1249-0},
}
@InProceedings {civemsa22_all,
author = {A. Genovese},
booktitle = {Proc. of the 2022 IEEE Int. Conf. on Computational Intelligence and Virtual Environments for
Measurement Systems and Applications (CIVEMSA 2022)},
title = {ALLNet: Acute Lymphoblastic Leukemia detection using lightweight convolutional networks},
address = {Chemnitz, Germany},
pages = {1-6},
month = {June},
day = {15-17},
year = {2022},
note = {978-1-6654-3445-4}
}
@InProceedings {aimia23,
author = {A. Genovese and V. Piuri and F. Scotti},
booktitle = {Proc. of the IEEE Int. Conf. on Acoustics Speech and Signal Processing Workshops
(ICASSPW 2023)},
title = {ALL-IDB Patches: Whole slide imaging for Acute Lymphoblastic Leukemia detection using
Deep Learning},
address = {Rhodes Island, Greece},
pages = {1-5},
month = {June},
day = {4-10},
year = {2023},
note = {979-8-3503-0261-5}
}
@Article {access23,
author = {A. Genovese and V. Piuri and K. N. Plataniotis and F. Scotti},
title = {DL4ALL: Multi-task cross-dataset transfer learning for Acute Lymphoblastic Leukemia detection},
journal = {IEEE Access},
volume = {11},
pages = {65222-65237},
year = {2023},
note = {2169-3536}
}
@Article {imavis24,
author = {A. Genovese and V. Piuri and F. Scotti},
title = {A Decision Support System for Acute Lymphoblastic Leukemia Detection based on Explainable Artificial Intelligence},
journal = {Image and Vision Computing},
volume = {151},
number = {105298},
month = {November},
year = {2024},
note = {0262-8856}
}
Acknowledgements
We gratefully acknowledge the support of NVIDIA Corporation with the donation of the Titan X Pascal GPU used for this research, within the project "Deep Learning and CUDA for advanced and less-constrained biometric systems".
People
- Angelo Genovese (proposer and maintainer), angelo.genovese AT unimi.it
- Mahdi S. Hosseini
- Vincenzo Piuri
- Konstantinos N. Plataniotis
- Fabio Scotti
Downloads
- Acute Lymphoblastic Leukemia detection based on adaptive unsharpening and Deep Learning: [GitHub]
- Histopathological transfer learning for Acute Lymphoblastic Leukemia detection: [GitHub]
- ALLNet: Acute Lymphoblastic Leukemia detection using lightweight convolutional networks: [GitHub]
- ALL-IDB Patches: Whole slide imaging for Acute Lymphoblastic Leukemia detection using Deep Learning: [GitHub]
- DL4ALL: Multi-task cross-dataset transfer learning for Acute Lymphoblastic Leukemia detection: [GitHub]
- A decision support system for Acute Lymphoblastic Leukemia detection based on Explainable Artificial Intelligence: [GitHub]
Related projects
- Acute Lymphoblastic Leukemia Image Database for Image Processing: ALL-IDB


